Matching Sites for Future Students
- Name
- KOBAYASHI, Kappei
- Position
- Professor
- Department
- Food Production Science
- Course
- Agro-Biological Science
- Field of study
- Molecular bio-resources
- Mail address
- kappei@ehime-u.ac.jp
- Research subject
- Exploration and identification of plant virus pathogens, and development of diagnostic methods
- Keyword
- Plant virus
Comprehensive search
Novel virus
Genetic diagnosis
Nucleic acid extraction
- Research content
-
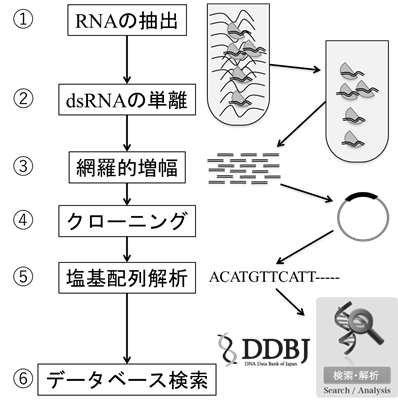
①RNA extraction
②Isolation of dsRNA
③Comprehensively amplified
④Cloning
⑤Analysis of base sequences
⑥Data base searching
We quickly find candidate causative pathogens of plant diseases suspected by virus, and develop a simple and rapid gene diagnosis method.
Comprehensive plant virus detection method
We can catch the tail of the virus even from plants that do not know what virus is infecting
① First, RNA is extracted from RNA viruses that account for the majority of plant viruses.
② Concentrate double-stranded RNA (dsRNA) that plants do not make by themselves.
③ cDNA is synthesized from dsRNA, and all of it is comprehensively amplified.
④ Clone the DNA randomly amplified.
⑤Determine the base sequence of the randomly selected clone.
⑥ For all the determined nucleotide sequences, find out similar sequences from the viral gene sequences in the database.
Currently, ④ and ⑤ can be done in one step by using a Next Generation Sequencer. Take the virus body from the tail caught by this method, develop and provide genetic diagnostic methods such as PCR.
Kobayashi K, Tomita R and Sakamoto M. (2009) Journal of General Plant Pathology. 75, 87-91(Received JCPP Paper award 2011).
Yanagisawa H, et. Al. (other 5 co-authors)( Kobayashi K and Sekine K.-T. (2016) Viruses 8(3): 70(Use a Next Generation Sequencer)
Achievement of new virus discovery
Gentian bull virus associated virus: Kobayashi, K. et.al. (2013) Journal of General Plant Pathology 79, 56-63.
Turkey goby ribbed virus: Shimomoto Y. et.al. (2014) Journal of General Plant Pathology 80, 169-175.
Simple and rapid plant nucleic acid extraction method for diagnosis of viral diseases
In order to perform genetic diagnostic methods such as PCR, it is necessary to extract nucleic acids firstly. We have developed a simple and rapid nucleic acid extraction method using filter paper and hammer. We are also working on improvements to concentrate viral nucleic acids.
① Paste a vinyl tape on the facing axis (front side) of a leaf.
② Put the leaf of ① into two sheets of filter papers, put it in a clear folder, hit with a hammer to destroy the tissue of the leaf, let the juice infiltrate to the filter papers.
③Remove leaf residue together with the vinyl tape.
④ Cut the filter paper sucking the crushed leaf juice to about 2 cm2 .
⑤ Round the paper filter piece of ④ into a cylindrical shape and place it in a micro centrifuge tube.
⑥ The plant component containing a nucleic acid is dissolved out into an extract containing a strong surfactant, then extract the nucleic acid by the usual method.Akhter MS et.al.(2016)Journal of General Plant Pathology 82: 268-272.
- Resources, technologies, other available resources
- Information of pathogen candidate virus gene extracted by comprehensive search method from diseased plants whose causative pathogen is unknown. Genetic diagnosis technology (nucleic acid extraction method, design of PCR primers)
- Project research theme
- Identification of a causative pathogenic virus in new diseases of important crops
Development of convenient, rapid and in-expensive nucleic acid extraction method for diagnosis of viral diseases
- Study Topics
- Others
